generate_sbm#
- graph_tool.generation.generate_sbm(b, probs, out_degs=None, in_degs=None, directed=False, micro_ers=False, micro_degs=False, condensed=False)[source]#
Generate a random graph by sampling from the Poisson or microcanonical stochastic block model.
- Parameters:
- biterable or
numpy.ndarray Group membership for each vertex.
- probstwo-dimensional
numpy.ndarrayorscipy.sparse.spmatrix Matrix with edge propensities between groups. The value
probs[r,s]corresponds to the average number of edges between groupsrands(or twice the average number ifr == sand the graph is undirected).- out_degsiterable or
numpy.ndarray(optional, default:None) Out-degree propensity for each vertex. If not provided, a constant value will be used. Note that the values will be normalized inside each group, if they are not already so.
- in_degsiterable or
numpy.ndarray(optional, default:None) In-degree propensity for each vertex. If not provided, a constant value will be used. Note that the values will be normalized inside each group, if they are not already so.
- directed
bool(optional, default:False) Whether the graph is directed.
- micro_ers
bool(optional, default:False) If
True, the microcanonical version of the model will be evoked, where the numbers of edges between groups will be given exactly by the parameterprobs, and this will not fluctuate between samples.- micro_degs
bool(optional, default:False) If
True, the microcanonical version of the degree-corrected model will be evoked, where the degrees of vertices will be given exactly by the parametersout_degsandin_degs, and they will not fluctuate between samples. (Ifmicro_degs == Trueit impliesmicro_ers == True.)- condensed
bool(optional, default:False) If
True, the graph returned will be a simple graph, with the edge multiplicites returned as a property map.
- biterable or
- Returns:
- g
Graph The generated graph.
- w
EdgePropertyMap(only ifcondensed == True) Edge property map with edge multiplicities.
- g
See also
random_graphrandom graph generation
Notes
The algorithm generates multigraphs with self-loops, according to the Poisson degree-corrected stochastic block model (SBM) [karrer-stochastic-2011], which includes the traditional SBM as a special case.
The multigraphs are generated with probability
\[P({\boldsymbol A}|{\boldsymbol \theta},{\boldsymbol \lambda},{\boldsymbol b}) = \prod_{i<j}\frac{e^{-\lambda_{b_ib_j}\theta_i\theta_j}(\lambda_{b_ib_j}\theta_i\theta_j)^{A_{ij}}}{A_{ij}!} \times\prod_i\frac{e^{-\lambda_{b_ib_i}\theta_i^2/2}(\lambda_{b_ib_i}\theta_i^2/2)^{A_{ij}/2}}{(A_{ij}/2)!},\]where \(\lambda_{rs}\) is the edge propensity between groups \(r\) and \(s\), and \(\theta_i\) is the propensity of vertex i to receive edges, which is proportional to its expected degree. Note that in the algorithm it is assumed that the vertex propensities are normalized for each group,
\[\sum_i\theta_i\delta_{b_i,r} = 1,\]such that the value \(\lambda_{rs}\) will correspond to the average number of edges between groups \(r\) and \(s\) (or twice that if \(r = s\)). If the supplied values of \(\theta_i\) are not normalized as above, they will be normalized prior to the generation of the graph.
For directed graphs, the probability is analogous, with \(\lambda_{rs}\) being in general asymmetric:
\[P({\boldsymbol A}|{\boldsymbol \theta},{\boldsymbol \lambda},{\boldsymbol b}) = \prod_{ij}\frac{e^{-\lambda_{b_ib_j}\theta^+_i\theta^-_j}(\lambda_{b_ib_j}\theta^+_i\theta^-_j)^{A_{ij}}}{A_{ij}!}.\]Again, the same normalization is assumed:
\[\sum_i\theta_i^+\delta_{b_i,r} = \sum_i\theta_i^-\delta_{b_i,r} = 1,\]such that the value \(\lambda_{rs}\) will correspond to the average number of directed edges between groups \(r\) and \(s\).
The traditional (i.e. non-degree-corrected) SBM is recovered from the above model by setting \(\theta_i=1/n_{b_i}\) (or \(\theta^+_i=\theta^-_i=1/n_{b_i}\) in the directed case), which is done automatically if
out_degsandin_degsare not specified.In case the parameter
micro_degs == Trueis passed, a microcanical model is used instead, where both the number of edges between groups as well as the degrees of the vertices are preserved exactly, instead of only on expectation [peixoto-nonparametric-2017]. In this case, the parameters are interpreted as \({\boldsymbol\lambda}\equiv{\boldsymbol e}\) and \({\boldsymbol\theta}\equiv{\boldsymbol k}\), where \(e_{rs}\) is the number of edges between groups \(r\) and \(s\) (or twice that if \(r=s\) in the undirected case), and \(k_i\) is the degree of vertex \(i\). This model is a generalization of the configuration model, where multigraphs are sampled with probability\[P({\boldsymbol A}|{\boldsymbol k},{\boldsymbol e},{\boldsymbol b}) = \frac{\prod_{r<s}e_{rs}!\prod_re_{rr}!!\prod_ik_i!}{\prod_re_r!\prod_{i<j}A_{ij}!\prod_iA_{ii}!!}.\]and in the directed case with probability
\[P({\boldsymbol A}|{\boldsymbol k}^+,{\boldsymbol k}^-,{\boldsymbol e},{\boldsymbol b}) = \frac{\prod_{rs}e_{rs}!\prod_ik^+_i!k^-_i!}{\prod_re^+_r!e^-_r!\prod_{ij}A_{ij}!}.\]where \(e^+_r = \sum_se_{rs}\), \(e^-_r = \sum_se_{sr}\), \(k^+_i = \sum_jA_{ij}\) and \(k^-_i = \sum_jA_{ji}\).
In the non-degree-corrected case, if
micro_ers == True, the microcanonical model corresponds to\[P({\boldsymbol A}|{\boldsymbol e},{\boldsymbol b}) = \frac{\prod_{r<s}e_{rs}!\prod_re_{rr}!!}{\prod_rn_r^{e_r}\prod_{i<j}A_{ij}!\prod_iA_{ii}!!},\]and in the directed case to
\[P({\boldsymbol A}|{\boldsymbol e},{\boldsymbol b}) = \frac{\prod_{rs}e_{rs}!}{\prod_rn_r^{e_r^+ + e_r^-}\prod_{ij}A_{ij}!}.\]In every case above, the final graph is generated in time \(O(V + E + B)\), where \(B\) is the number of groups.
References
[karrer-stochastic-2011]Brian Karrer and M. E. J. Newman, “Stochastic blockmodels and community structure in networks,” Physical Review E 83, no. 1: 016107 (2011) DOI: 10.1103/PhysRevE.83.016107 [sci-hub, @tor] arXiv: 1008.3926
[peixoto-nonparametric-2017]Tiago P. Peixoto, “Nonparametric Bayesian inference of the microcanonical stochastic block model”, Phys. Rev. E 95 012317 (2017). DOI: 10.1103/PhysRevE.95.012317 [sci-hub, @tor], arXiv: 1610.02703
Examples
>>> g = gt.collection.data["polblogs"] >>> g = gt.GraphView(g, vfilt=gt.label_largest_component(g)) >>> g = gt.Graph(g, prune=True) >>> state = gt.minimize_blockmodel_dl(g) >>> u = gt.generate_sbm(state.b.a, gt.adjacency(state.get_bg(), ... state.get_ers()).T, ... g.degree_property_map("out").a, ... g.degree_property_map("in").a, directed=True) >>> gt.graph_draw(g, g.vp.pos, output="polblogs-sbm.pdf") <...> >>> gt.graph_draw(u, u.own_property(g.vp.pos), output="polblogs-sbm-generated.pdf") <...>
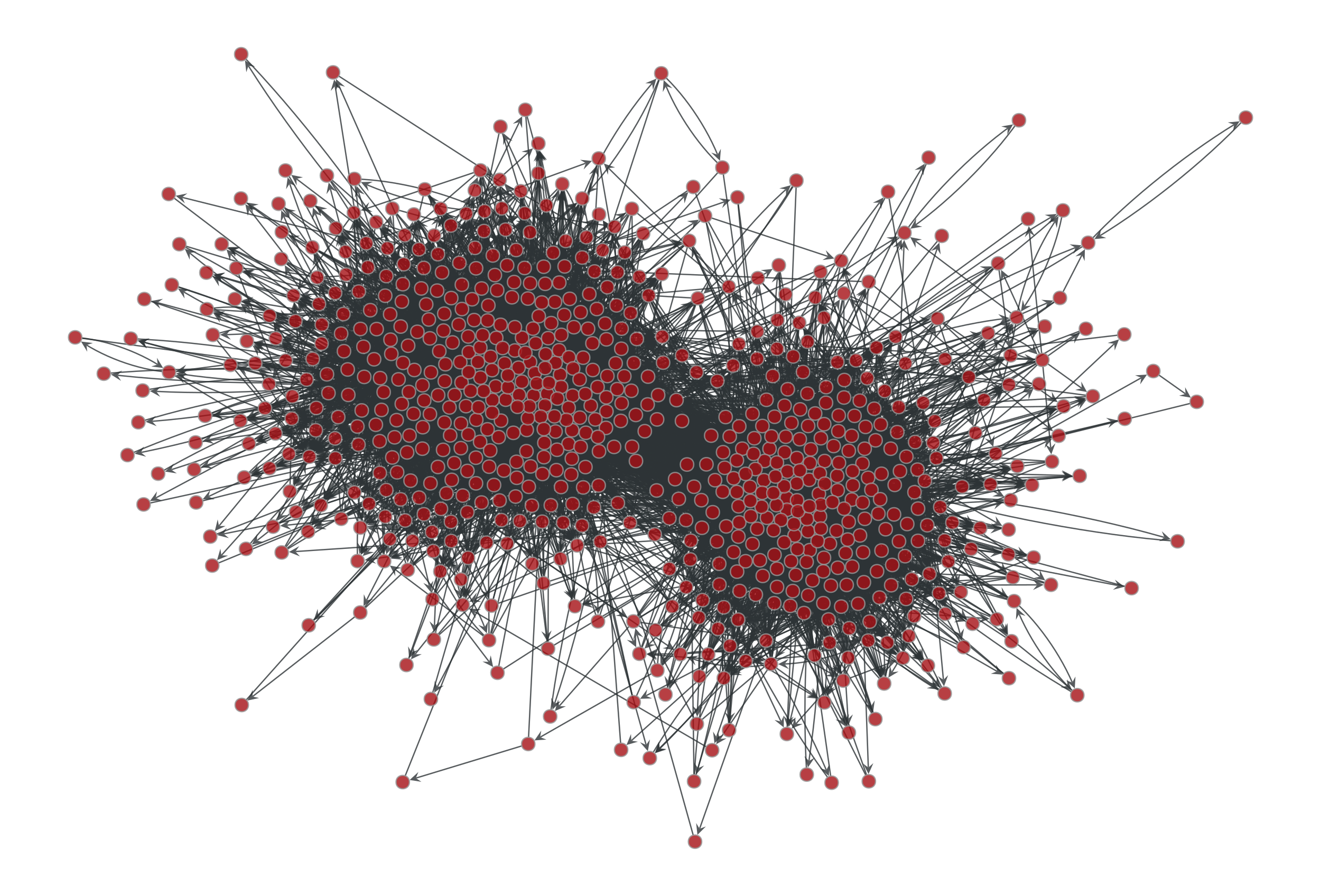
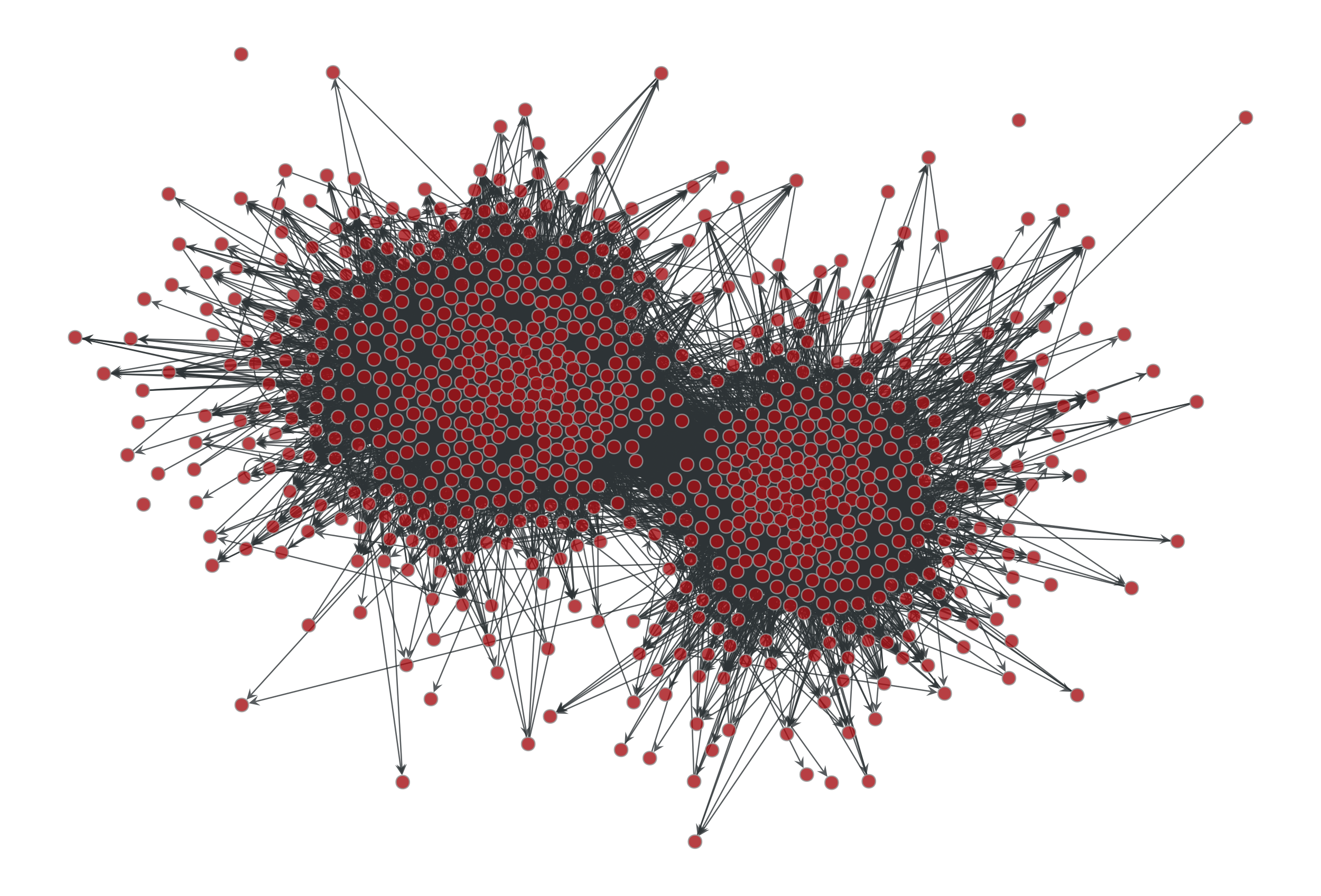
Left: Political blogs network. Right: Sample from the degree-corrected SBM fitted to the original network.